Enhancer methylation dynamics drive core transcriptional regulatory circuitry in pan-cancer
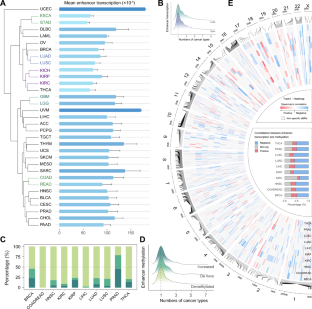
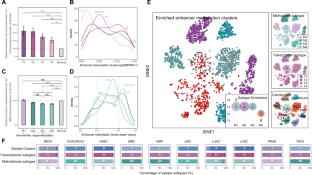
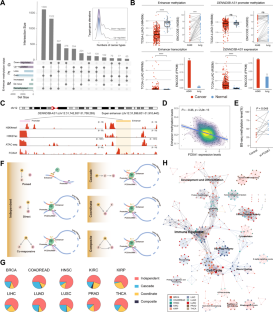
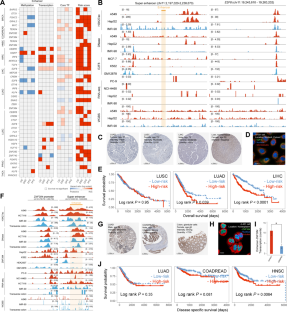
Accumulating evidence has demonstrated that enhancer methylation has strong and dynamic regulatory effects on gene expression. Some transcription factors (TFs) can auto- and cross-regulate in a feed-forward manner, and cooperate with their enhancers to form core transcriptional regulatory circuitries (CRCs). However, the elaborated regulatory mechanism between enhancer methylation and CRC remains the tip of the iceberg. Here, we revealed that DNA methylation could drive the tissue-specific enhancer basal transcription and target gene expression in human cancers. By integrating methylome, transcriptome, and 3D genomic data, we identified enhancer methylation triplets (enhancer methylation-enhancer transcription-target gene expression) and dissected potential regulatory patterns within them. Moreover, we observed that cancer-specific core TFs regulated by enhancers were able to shape their enhancer methylation forming the enhancer methylation-driven CRCs (emCRCs). Further parsing of clinical implications showed rewired emCRCs could serve as druggable targets and prognostic risk markers. In summary, the integrative analysis of enhancer methylation regulome would facilitate portraying the cancer epigenomics landscape and developing the epigenetic anti-cancer approaches.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
265,23 € per year
only 5,30 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Epigenomic landscape of human colorectal cancer unveils an aberrant core of pan-cancer enhancers orchestrated by YAP/TAZ
Article Open access 20 April 2021
Integrative pan cancer analysis reveals epigenomic variation in cancer type and cell specific chromatin domains
Article Open access 03 March 2021
MeinteR: A framework to prioritize DNA methylation aberrations based on conformational and cis-regulatory element enrichment
Article Open access 16 December 2019
Data availability
All the data used in the analysis can be obtained from TCGA, ENCODE, and GEO.
Code availability
The data analysis in this manuscript was conducted using custom scripts, all available at http://bio-bigdata.tech/EmethCRC.
References
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. ArticleCASPubMedGoogle Scholar
- Hegde AN, Smith SG. Recent developments in transcriptional and translational regulation underlying long-term synaptic plasticity and memory. Learn Mem. 2019;26:307–17. ArticleCASPubMedPubMed CentralGoogle Scholar
- Kim S, Kaang B-K. Epigenetic regulation and chromatin remodeling in learning and memory. Exp Mol Med. 2017;49:e281–e281. ArticlePubMedPubMed CentralCASGoogle Scholar
- Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. ArticleCASPubMedGoogle Scholar
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. ArticleCASPubMedPubMed CentralGoogle Scholar
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–9. ArticleCASPubMedGoogle Scholar
- Schuijers J, Manteiga JC, Weintraub AS, Day DS, Zamudio AV, Hnisz D, et al. Transcriptional dysregulation of MYC reveals common enhancer-docking mechanism. Cell Rep. 2018;23:349–60. ArticleCASPubMedPubMed CentralGoogle Scholar
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–84. ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet. 2016;17:551–65. ArticleCASPubMedPubMed CentralGoogle Scholar
- Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schübeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–9. ArticleCASPubMedGoogle Scholar
- Sato N, Kondo M, Arai K. The orphan nuclear receptor GCNF recruits DNA methyltransferase for Oct-3/4 silencing. Biochem Biophys Res Commun. 2006;344:845–51. ArticleCASPubMedGoogle Scholar
- Barnett KR, Decato BE, Scott TJ, Hansen TJ, Chen B, Attalla J, et al. ATAC-Me captures prolonged DNA methylation of dynamic chromatin accessibility loci during cell fate transitions. Mol Cell. 2020;77:1350–.e1356. ArticleCASPubMedPubMed CentralGoogle Scholar
- Vanzan L, Soldati H, Ythier V, Anand S, Braun SMG, Francis N, et al. High throughput screening identifies SOX2 as a super pioneer factor that inhibits DNA methylation maintenance at its binding sites. Nat Commun. 2021;12:3337. ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhang Y, Zhang D, Li Q, Liang J, Sun L, Yi X, et al. Nucleation of DNA repair factors by FOXA1 links DNA demethylation to transcriptional pioneering. Nat Genet. 2016;48:1003–13. ArticleCASPubMedGoogle Scholar
- Saint-André V, Federation AJ, Lin CY, Abraham BJ, Reddy J, Lee TI, et al. Models of human core transcriptional regulatory circuitries. Genome Res. 2016;26:385–96. ArticlePubMedPubMed CentralCASGoogle Scholar
- Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22:209–21. ArticleCASPubMedPubMed CentralGoogle Scholar
- Decaesteker B, Denecker G, Van Neste C, Dolman EM, Van Loocke W, Gartlgruber M, et al. TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets. Nat Commun. 2018;9:4866. ArticlePubMedPubMed CentralCASGoogle Scholar
- Xu Q, Chen J, Ni S, Tan C, Xu M, Dong L, et al. Pan-cancer transcriptome analysis reveals a gene expression signature for the identification of tumor tissue origin. Mod Pathol. 2016;29:546–56. ArticlePubMedCASGoogle Scholar
- Chen H, Li C, Peng X, Zhou Z, Weinstein JN. Cancer Genome Atlas Research N, et al. A pan-cancer analysis of enhancer expression in nearly 9000 patient samples. Cell. 2018;173:386–99 e312. ArticleCASPubMedPubMed CentralGoogle Scholar
- Petell CJ, Alabdi L, He M, San Miguel P, Rose R, Gowher H. An epigenetic switch regulates de novo DNA methylation at a subset of pluripotency gene enhancers during embryonic stem cell differentiation. Nucleic Acids Res. 2016;44:7605–17. ArticleCASPubMedPubMed CentralGoogle Scholar
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–91. ArticleCASPubMedPubMed CentralGoogle Scholar
- Wiench M, John S, Baek S, Johnson TA, Sung MH, Escobar T, et al. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30:3028–39. ArticleCASPubMedPubMed CentralGoogle Scholar
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–83. ArticleCASPubMedGoogle Scholar
- Benner C, Isoda T, Murre C. New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors. Proc Natl Acad Sci USA. 2015;112:12776–81. ArticleCASPubMedPubMed CentralGoogle Scholar
- Bell RE, Golan T, Sheinboim D, Malcov H, Amar D, Salamon A, et al. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 2016;26:601–11. ArticleCASPubMedPubMed CentralGoogle Scholar
- Song Y, van den Berg PR, Markoulaki S, Soldner F, Dall’Agnese A, Henninger JE, et al. Dynamic Enhancer DNA methylation as basis for transcriptional and cellular heterogeneity of ESCs. Mol Cell. 2019;75:905–20 e906. ArticleCASPubMedPubMed CentralGoogle Scholar
- Paralkar Vikram R, Taborda Cristian C, Huang P, Yao Y, Kossenkov Andrew V, Prasad R, et al. Unlinking an lncRNA from its associated cis element. Mol Cell. 2016;62:104–10. ArticleCASPubMedPubMed CentralGoogle Scholar
- Aguilo F, Li S, Balasubramaniyan N, Sancho A, Benko S, Zhang F, et al. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1α. Cell Rep. 2016;14:479–92. ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhao Y, Wang L, Ren S, Wang L, Blackburn PR, McNulty MS, et al. Activation of P-TEFb by androgen receptor-regulated enhancer RNAs in castration-resistant prostate cancer. Cell Rep. 2016;15:599–610. ArticleCASPubMedPubMed CentralGoogle Scholar
- Miao Y, Ajami NE, Huang T-S, Lin F-M, Lou C-H, Wang Y-T, et al. Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat Commun. 2018;9:292. ArticlePubMedPubMed CentralCASGoogle Scholar
- Xiong L, Wu F, Wu Q, Xu L, Cheung OK, Kang W, et al. Aberrant enhancer hypomethylation contributes to hepatic carcinogenesis through global transcriptional reprogramming. Nat Commun. 2019;10:335. ArticlePubMedPubMed CentralCASGoogle Scholar
- Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. ArticlePubMedPubMed CentralCASGoogle Scholar
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–7. ArticleCASPubMedPubMed CentralGoogle Scholar
- Tsamardinos I, Borboudakis G. Permutation Testing Improves Bayesian Network Learning. In: Balcázar JL, Bonchi F, Gionis A, Sebag M (eds). Machine Learning and Knowledge Discovery in Databases. Springer Berlin Heidelberg: Berlin, Heidelberg, 2010, pp 322–37. https://doi.org/10.1007/978-3-642-15939-8_21.
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. ArticlePubMedPubMed CentralCASGoogle Scholar
- Suzuki T, Maeda S, Furuhata E, Shimizu Y, Nishimura H, Kishima M, et al. A screening system to identify transcription factors that induce binding site-directed DNA demethylation. Epigenetics Chromatin. 2017;10:60. ArticlePubMedPubMed CentralCASGoogle Scholar
- Feldmann A, Ivanek R, Murr R, Gaidatzis D, Burger L, Schübeler D. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet. 2013;9:e1003994–e1003994. ArticlePubMedPubMed CentralCASGoogle Scholar
- Xiong L, Wu F, Wu Q, Xu L, Cheung OK, Kang W, et al. Aberrant enhancer hypomethylation contributes to hepatic carcinogenesis through global transcriptional reprogramming. Nat Commun. 2019;10:335. ArticlePubMedPubMed CentralCASGoogle Scholar
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. ArticleCASPubMedPubMed CentralGoogle Scholar
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem. Cells Cell. 2005;122:947–56. ArticleCASPubMedGoogle Scholar
- Chung VY, Tan TZ, Ye J, Huang R-L, Lai H-C, Kappei D, et al. The role of GRHL2 and epigenetic remodeling in epithelial–mesenchymal plasticity in ovarian cancer cells. Commun Biol. 2019;2:272. ArticlePubMedPubMed CentralCASGoogle Scholar
- Baek S-J, Kim M, Bae D-H, Kim J-H, Kim H-J, Han M-E, et al. Integrated epigenomic analyses of enhancer as well as promoter regions in gastric cancer. Oncotarget. 2016;7:25620–31. ArticlePubMedPubMed CentralGoogle Scholar
- Home P, Saha B, Ray S, Dutta D, Gunewardena S, Yoo B, et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci USA. 2012;109:7362–7. ArticleCASPubMedPubMed CentralGoogle Scholar
- Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–d386. ArticleCASPubMedGoogle Scholar
- Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. ArticlePubMedPubMed CentralCASGoogle Scholar
- Sladky VC, Knapp K, Soratroi C, Heppke J, Eichin F, Rocamora-Reverte L, et al. E2F-family members engage the PIDDosome to limit hepatocyte ploidy in liver development and regeneration. Dev Cell. 2020;52:335–.e337. ArticleCASPubMedGoogle Scholar
- Kent LN, Rakijas JB, Pandit SK, Westendorp B, Chen H-Z, Huntington JT, et al. E2f8 mediates tumor suppression in postnatal liver development. J Clin Investig. 2016;126:2955–69. ArticlePubMedPubMed CentralGoogle Scholar
- Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer. 2018;18:696–705. ArticleCASPubMedPubMed CentralGoogle Scholar
- Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46:D1068–D1073. ArticleCASPubMedGoogle Scholar
- Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;1:1–16. Google Scholar
- Priness I, Maimon O, Ben-Gal I. Evaluation of gene-expression clustering via mutual information distance measure. BMC Bioinform. 2007;8:111. ArticleCASGoogle Scholar
- Steuer R, Kurths J, Daub CO, Weise J, Selbig J. The mutual information: detecting and evaluating dependencies between variables. Bioinformatics. 2002;18:S231–S240. ArticlePubMedGoogle Scholar
- Niculescu RS, Mitchell TM, Rao RB, Bennett KP, Parrado-Hernández E. Bayesian network learning with parameter constraints. J Mach Learn Res. 2006;7:1357–83. Google Scholar
- Qian M, Zhang H, Kham SK, Liu S, Jiang C, Zhao X, et al. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res. 2017;27:185–95. ArticleCASPubMedPubMed CentralGoogle Scholar
- Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013;4:578–88. ArticleCASPubMedGoogle Scholar
- Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75:3728–37. ArticleCASPubMedPubMed CentralGoogle Scholar
- Tong Y, Sun J, Wong CF, Kang Q, Ru B, Wong CN, et al. MICMIC: identification of DNA methylation of distal regulatory regions with causal effects on tumorigenesis. Genome Biol. 2018;19:73. ArticlePubMedPubMed CentralCASGoogle Scholar
- Wagenmakers E-J, Farrell S. AIC model selection using Akaike weights. Psychonomic Bull Rev. 2004;11:192–6. ArticleGoogle Scholar
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–D801. ArticleCASPubMedGoogle Scholar
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. ArticleCASPubMedPubMed CentralGoogle Scholar
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. ArticleCASPubMedPubMed CentralGoogle Scholar
- Mei S, Meyer CA, Zheng R, Qin Q, Wu Q, Jiang P, et al. Cistrome cancer: a web resource for integrative gene regulation modeling in cancer. Cancer Res. 2017;77:e19–e22. ArticleCASPubMedPubMed CentralGoogle Scholar
- Yao L, Shen H, Laird PW, Farnham PJ, Berman BP. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol. 2015;16:105. ArticlePubMedPubMed CentralCASGoogle Scholar
Acknowledgements
The authors gratefully thank the TCGA, ENCODE, TANRIC, and GEO for providing data for this work.









